Special Issue in Virology: Functional Genomics of Papillomaviruses

Special issue: The Papillomavirus Episteme
Paul F. Lamberta, Alison McBrideb and Hans Ulrich Bernardc
a University of Wisconsin, USA
b National Institutes of Health, USA
c University of California, USA
Papillomaviruses are an ancient group of viruses that have coevolved along with their host species for millions of years; each virus is species specific and trophic for specific anatomical niches in the host epithelium. Papillomavirus infections can be asymptomatic, or can give rise to a wide range of pathologies, and a subset of papillomaviruses is strongly associated with human cancer. Unlike many other viruses, papillomaviruses are genetically stable and evolve at a slow rate along with the host. The availability of complete genome sequences from hundreds of different papillomaviruses provides a treasure trove of information for comparative genomics. This Special Issue is intended to be a companion resource to the Papillomavirus Episteme bioinformatics website (http://pave.niaid.nih.gov). The Papillomavirus Episteme hosts organized and curated information related to viral genomics. Each article in the Special Issue: Functional Genomics of Papillomaviruses provides an in-depth, unbiased and encyclopedic overview of different aspects of papillomavirus genomics and proteomics, written by experts in the papillomavirus field. Unlike other reviews, the main aim of each article is to completely document published details about each topic, so that this Special Issue will serve as an important reference resource for the scientific community. You can visit the blog site for this special issue at http://www.virologyhighlights.com/?p=144.
Cross-roads in the classification of papillomaviruses
Ethel-Michele de Villiers
Deutsches Krebsforschungszentrum, Germany
Acceptance of an official classification for the family Papillomaviridae based purely on DNA sequence relatedness, was achieved as late as 2003. The rate of isolation and characterization of new papillomavirus types has greatly depended on and subjected to the development of new laboratory techniques. Introduction of every new technique led to a temporarily burst in the number of new isolates. In the following, the bumpy road towards achieving a classification system combined with the controversies of implementing and accepting new techniques will be summarized. An update of the classification of the 170 human papillomavirus (HPV) types presently known is presented. Arguments towards the implementation of metagenomic sequencing for this rapidly growing family will be presented.
Evolution of the Papillomaviridae
Koenraad Van Doorslaer
National Institutes of Health, USA
Viruses belonging to the Papillomaviridae family have been isolated from a variety of mammals, birds and non-avian reptiles. It is likely that most, if not all, amniotes carry a broad array of viral types. To date, the complete genomic sequence of more than 240 distinct viral types has been characterized at the nucleotide level. The analysis of this sequence information has begun to shed light on the evolutionary history of this important virus family. The available data suggests that many different evolutionary mechanisms have influenced the papillomavirus phylogenetic tree. Increasing evidence supports that the ancestral papillomavirus initially specialized to infect different ecological niches on the host. This episode of niche sorting was followed by extensive episodes of co-speciation with the host. This review attempts to summarize our current understanding of the papillomavirus evolution.
Diseases associated with human papillomavirus infection
Heather A. Cubie
University of Edinburgh, UK
Human papillomaviruses (HPVs) are ubiquitous, well adapted to their host and cleverly sequestered away from immune responses. HPV infections can be productive, subclinical or latent in both skin and mucosa. The causal association of HPV with cervical cancer, and increasingly with rising numbers of squamous cell carcinomas at other sites in both men and women, is increasingly recognised, while the morbidity of cutaneous HPV lesions, particularly in the immunosuppressed population is also significant. This chapter sets out the range of infections and clinical manifestations of the consequences of infection and its persistence and describes why HPVs are both highly effective pathogens and carcinogens, challenging to eliminate.
Monika Bergvalla, Thomas Melendyb and Jacques Archambaulta
a McGill University, Canada
b University of Buffalo, USA
E1, an ATP-dependent DNA helicase, is the only enzyme encoded by papillomaviruses (PVs). It is essential for replication and amplification of the viral episome in the nucleus of infected cells. To do so, E1 assembles into a double-hexamer at the viral origin, unwinds DNA at the origin and ahead of the replication fork and interacts with cellular DNA replication factors. Biochemical and structural studies have revealed the assembly pathway of E1 at the origin and how the enzyme unwinds DNA using a spiral escalator mechanism. E1 is tightly regulated in vivo, in particular by post-translational modifications that restrict its accumulation in the nucleus. Here we review how different functional domains of E1 orchestrate viral DNA replication, with an emphasis on their interactions with substrate DNA, host DNA replication factors and modifying enzymes. These studies have made E1 one of the best characterized helicases and provided unique insights on how PVs usurp different host-cell machineries to replicate and amplify their genome in a tightly controlled manner.
Movies of DNA translocation in the E1 replicative hexameric helicase
The Papillomavirus E2 proteins
Alison A. McBride
National Institutes of Health, USA
The papillomavirus E2 proteins are pivotal to the viral life cycle and have well characterized functions in transcriptional regulation, initiation of DNA replication and partitioning the viral genome. The E2 proteins also function in vegetative DNA replication, post-transcriptional processes and possibly packaging. This review describes structural and functional aspects of the E2 proteins and their binding sites on the viral genome. It is intended to be a reference guide to this viral protein.
The E4 protein; structure, function and patterns of expression
John Doorbar
National Institute of Medical Research, UK
The papillomavirus E4 open reading frame (ORF) is contained within the E2 ORF, with the primary E4 gene-product (E1∧E4) being translated from a spliced mRNA that includes the E1 initiation codon and adjacent sequences. E4 is located centrally within the E2 gene, in a region that encodes the E2 protein′s flexible hinge domain. Although a number of minor E4 transcripts have been reported, it is the product of the abundant E1∧E4 mRNA that has been most extensively analysed.
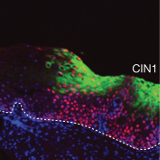
During the papillomavirus life cycle, the E1∧E4 gene products generally become detectable at the onset of vegetative viral genome amplification as the late stages of infection begin. E4 contributes to genome amplification success and virus synthesis, with its high level of expression suggesting additional roles in virus release and/or transmission. In general, E4 is easily visualised in biopsy material by immunostaining, and can be detected in lesions caused by diverse papillomavirus types, including those of dogs, rabbits and cattle as well as humans. The E4 protein can serve as a biomarker of active virus infection, and in the case of high-risk human types also disease severity. In some cutaneous lesions, E4 can be expressed at higher levels than the virion coat proteins, and can account for as much as 30% of total lesional protein content. The E4 proteins of the Beta, Gamma and Mu HPV types assemble into distinctive cytoplasmic, and sometimes nuclear, inclusion granules.
In general, the E4 proteins are expressed before L2 and L1, with their structure and function being modified, first by kinases as the infected cell progresses through the S and G2 cell cycle phases, but also by proteases as the cell exits the cell cycle and undergoes true terminal differentiation. The kinases that regulate E4 also affect other viral proteins simultaneously, and include protein kinase A, Cyclin-dependent kinase, members of the MAP Kinase family and protein kinase C. For HPV16 E1∧E4, these kinases regulate one of the E1∧E4 proteins main functions, the association with the cellular keratin network, and eventually also its cleavage by the protease calpain which allows assembly into amyloid-like fibres and reorganisation of the keratin network. Although the E4 proteins of different HPV types appear divergent at the level of their primary amino acid sequence, they share a recognisable modular organisation and pattern of expression, which may underlie conserved functions and regulation. Assembly into higher-order multimers and suppression of cell proliferation are common to all E4 proteins examined. Although not yet formally demonstrated, a role in virus release and transmission remains a likely function for E4.
Daniel DiMaio and Lisa M. Petti
Yale University, USA
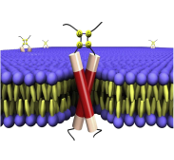
The E5 proteins are short transmembrane proteins encoded by many animal and human papillomaviruses. These proteins display transforming activity in cultured cells and animals, and they presumably also play a role in the productive virus life cycle. The E5 proteins are thought to act by modulating the activity of cellular proteins. Here, we describe the biological activities of the best-studied E5 proteins and discuss the evidence implicating specific protein targets and pathways in mediating these activities. The primary target of the 44-amino acid BPV1 E5 protein is the PDGF β receptor, whereas the EGF receptor appears to be an important target of the 83-amino acid HPV16 E5 protein. Both E5 proteins also bind to the vacuolar ATPase and affect MHC class I expression and cell–cell communication. Continued studies of the E5 proteins will elucidate important aspects of transmembrane protein–protein interactions, cellular signal transduction, cell biology, virus replication, and tumorigenesis.
Papillomavirus E6 oncoproteins
Scott B. Vande Pola and Aloysius J. Klingelhutzb
a University of Virginia, USA
b University of Iowa, USA
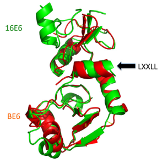
Papillomaviruses induce benign and malignant epithelial tumors, and the viral E6 oncoprotein is essential for full transformation. E6 contributes to transformation by associating with cellular proteins, docking on specific acidic LXXLL peptide motifs found on these proteins. This review examines insights from recent studies of human and animal E6 proteins that determine the three-dimensional structure of E6 when bound to acidic LXXLL peptides. The structure of E6 is related to recent advances in the purification and identification of E6 associated protein complexes. These E6 protein-complexes, together with other proteins that bind to E6, alter a broad array of biological outcomes including modulation of cell survival, cellular transcription, host cell differentiation, growth factor dependence, DNA damage responses, and cell cycle progression.
The papillomavirus E7 proteins
Ann Romana and Karl Mungerb
a Fred Hutchinson Cancer Research, USA
b Harvard Medical School, USA
E7 is an accessory protein that is not encoded by all papillomaviruses. The E7 amino terminus contains two regions of similarity to conserved regions 1 and 2 of the adenovirus E1A protein, which are also conserved in the simian vacuolating virus 40 large tumor antigen. The E7 carboxyl terminus consists of a zinc-binding motif, which is related to similar motifs in E6 proteins. E7 proteins play a central role in the human papillomavirus life cycle, reprogramming the cellular environment to be conducive to viral replication. E7 proteins encoded by the cancer-associated alpha human papillomaviruses have potent transforming activities, which together with E6, are necessary but not sufficient to render their host squamous epithelial cell tumorigenic. This article strives to provide a comprehensive summary of the published research studies on human papillomavirus E7 proteins.
The papillomavirus major capsid protein L1
Christopher B. Buck, Patricia M. Day and Benes L. Trus
National Institutes of Health, USA
The elegant icosahedral surface of the papillomavirus virion is formed by a single protein called L1. Recombinant L1 proteins can spontaneously self-assemble into a highly immunogenic structure that closely mimics the natural surface of native papillomavirus virions. This has served as the basis for two highly successful vaccines against cancer-causing human papillomaviruses (HPVs). During the viral life cycle, the capsid must undergo a variety of conformational changes, allowing key functions including the encapsidation of the ~8 kb viral genomic DNA, maturation into a more stable state to survive transit between hosts, mediating attachment to new host cells, and finally releasing the viral DNA into the newly infected host cell. This brief review focuses on conserved sequence and structural features that underlie the functions of this remarkable protein.
L2, the minor capsid protein of papillomavirus
Joshua W. Wanga and Richard B.S. Roden
The Johns Hopkins University, USA
We provide an overview of the host range, taxonomic classification and genomic diversity of animal papillomaviruses. The complete genomes of 112 non-human papillomavirus types, recovered from 54 different host species, are currently available in GenBank. The recent characterizations of reptilian papillomaviruses extend the host range of the Papillomaviridae to include all amniotes. Although the genetically diverse papillomaviruses have a highly conserved genomic lay-out, deviations from this prototypic genome organization are observed in several animal papillomaviruses, and only the core ORFs E1, E2, L2 and L1 are present in all characterized papillomavirus genomes. The discovery of papilloma–polyoma hybrids BPCV1 and BPCV2, containing a papillomaviral late region but an early region encoding typical polyomaviral nonstructural proteins, and the detection of recombination breakpoints between the early and late coding regions of cetacean papillomaviruses, could indicate that early and late gene cassettes of papillomaviruses are relatively independent entities that can be interchanged by recombination.
Papillomavirus transcripts and posttranscriptional regulation
Stefan Schwartz
Lund University, Sweden
Papillomavirus gene expression is strictly linked to the differentiation state of the infected cell and is highly regulated at the level of transcription and RNA processing. All papillomaviruses make extensive use of alternative mRNA polyadenylation and splicing to control gene expression. This chapter contains a compilation of all known alternatively spliced papillomavirus mRNAs and it summarizes our current knowledge of viral RNA elements, and viral and cellular factors that control papillomavirus mRNA processing.
Regulatory elements in the viral genome
Hans-Ulrich Bernard
University of California Irvine, United States
The small DNA genomes of papillomaviruses contain a surprisingly large number of regulatory or cis-responsive elements, which regulate replication and transcription of the virus, and control details like specificity for certain epithelial cells, specificity for layers in squamous epithelia, feedback mechanisms and coupling between host cell physiology and virus biology. Most of these elements occur in the long control region, while others are located elsewhere in the genome. Many papillomaviruses show a similar composition of cis-responsive elements, although these are scattered and do not occur as long segments of sequence similarity. This review summarizes our knowledge of the regulatory elements in several well-studied Alphapapillomavirus types, and indicates some similarities to other papillomavirus genera, whose properties are yet poorly understood.
Epigenetics of human papillomaviruses
Eric Johannsen and Paul F. Lambert
University of Wisconsin, USA
Human papilllomaviruses (HPVs) are common human pathogens that infect cutaneous or mucosal epithelia in which they cause warts, self-contained benign lesions that commonly regress. The HPV life cycle is intricately tied to the differentiation of the host epithelium it infects. Mucosotropic HPVs are the most common sexually transmitted pathogen known to mankind. A subset of the mucosotropic HPVs, so-called high risk HPVs, is etiologically associated with numerous cancers of the anogenital tract, most notably the cervix, as well as a growing fraction of head and neck cancers. In these cancers, the HPV genome, which normally exists an a double stranded, circular, nuclear plasmid, is commonly found integrated into the host genome and expresses two viral oncogenes, E6 and E7, that are implicated in the development and maintainance of the cancers caused by these high risk HPVs. Numerous studies, primarily on the high risk HPV16, have documented that the methylation status of the viral genome changes not only in the context of the viral life cycle but also in the context of the progressive neoplastic disease that culminates in cancer. In this article, we summarize the knowledge gained from those studies. We also provide the first analysis of available ChIP-seq data on the occupancy of both epigentically modified histones as well as transcription factors on the high risk HPV18 genome in the context of HeLa cells, a cervical cancer-derived cell line that has been the subject of extensive analyses using this technique.
Annabel Rector and Marc Van Ranst
University of Leuven, Belgium
We provide an overview of the host range, taxonomic classification and genomic diversity of animal papillomaviruses. The complete genomes of 112 non-human papillomavirus types, recovered from 54 different host species, are currently available in GenBank. The recent characterizations of reptilian papillomaviruses extend the host range of the Papillomaviridae to include all amniotes. Although the genetically diverse papillomaviruses have a highly conserved genomic lay-out, deviations from this prototypic genome organization are observed in several animal papillomaviruses, and only the core ORFs E1, E2, L2 and L1 are present in all characterized papillomavirus genomes. The discovery of papilloma–polyoma hybrids BPCV1 and BPCV2, containing a papillomaviral late region but an early region encoding typical polyomaviral nonstructural proteins, and the detection of recombination breakpoints between the early and late coding regions of cetacean papillomaviruses, could indicate that early and late gene cassettes of papillomaviruses are relatively independent entities that can be interchanged by recombination.
A systematic review of the prevalence of mucosal and cutaneous human papillomavirus types
Davit Bzhalavaa, Peng Guanb, Silvia Franceschib, Joakim Dillnera, b and Gary Cliffordb
a Karolinska Institutet, Sweden
b International Agency for Research on Cancer, France
Systematic reviews of the prevalence of different types of Human Papillomavirus (HPV) across a broad range of disease grades from normal to cancer are essential to gain basic knowledge of how widespread infections with the different HPV types are, and to provide information on the possible carcinogenicity of different HPV types. For HPV types that infect human mucosa, of which 12 are established causes of cervical cancer, we present the results of a systematic review and meta-analysis of 47 HPV types in cervical samples across the entire range of cervical diagnoses from normal to cervical cancer, restricted to studies using a number of well characterized PCR assays.
For the cutaneous HPV types, which have been linked to the development of squamous cell carcinoma of the skin, their presence has been measured in a variety of different sample types and by assays with variable performance. Therefore, we restricted a systematic review of their prevalence to studies that assayed for cutaneous HPV infection in a case-control format.
Human papillomavirus genome variants
Robert D. Burk, Ariana Harari and Zigui Chen
Albert Einstein College of Medicine, USA
Amongst the human papillomaviruses (HPVs), the genus Alphapapillomavirus contains HPV types that are uniquely pathogenic. They can be classified into species and types based on genetic distances between viral genomes. Current circulating infectious HPVs constitute a set of viral genomes that have evolved with the rapid expansion of the human population. Viral variants were initially identified through restriction enzyme polymorphisms and more recently through sequence determination of viral fragments. Using partial sequence information, the history of variants, and the association of HPV variants with disease will be discussed with the main focus on the recent utilization of full genome sequence information for variant analyses. The use of multiple sequence alignments of complete viral genomes and phylogenetic analyses have begun to define variant lineages and sublineages using empirically defined differences of 1.0–10.0% and 0.5–1.0%, respectively. These studies provide the basis to define the genetics of HPV pathogenesis.

