Papillomavirus capsids are composed of 72 capsomeres (pentamers) of the major capsid protein, L1. The L1 protein can self-assemble into viral like particles (VLPs) even in the absence of the minor capsid protein, L2, and VLPs are used to generate type-specific prophylactic vaccines. Although the L1 proteins are very well conserved among papillomaviruses, the surface loops are quite distinct between different viral types.
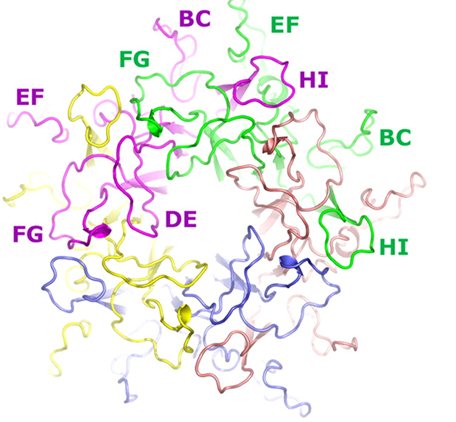
HPV16L1 pentamer. Each monomer is a different color and surface color and surface loops are labeled.
Reproduced with permission from Bishop B et al. J. Biol. Chem. 2007;282;31803-31811
L1 start site selection
The start position of most proteins on NCBI GenBank has been determined through the use of an 'open reading frame finder'. The problem with this approach is that it is unaware of biological evidence. In most cases these 'ORF finders' just look for the longest possible stretch of DNA located between a start and stop codon, thus ignoring the possible importance of downstream ATG codons.
For HPV16 it has been shown that L1 mRNA expression depends partly on derepression of late splice donor SD3632 and acceptor SA5639 (see review by Schwartz 2013). Importantly, multiple sequence alignments show that most PVs have a canonical splice acceptor immediately upstream of a completely conserved ATG.
However, in the case of some L1 proteins, an 'open reading frame finder' predicts an upstream ATG as the start codon (see figure below). At PaVE we tried to use as much available evidence as possible during the annotation process. Therefore, PaVE elected to take the SA5639 acceptor into account when annotating L1. This has the added advantage that all L1 sequences now have a well-defined and uniform start position.


